Faced with a dwindling number of active ingredients and a steady rise in fungicide resistance, growers are relying on plant breeders to produce new varieties able to cope with these modern challenges. CPM gets the gen on the research that will help breeders achieve this.
We’re providing breeders with signposts rather than precise directions.
By Lucy de la Pasture
Plant breeding takes a long time – finding the desired trait, breeding it in and then hoping it does what was intended. Then there’s a time lag while back-crosses are made to remove unwanted characteristics before a new variety is eventually born. Even with modern technology, the standard techniques are far from precise, explains Ellie Marshall, crop-breeding scientist at AHDB.
“AHDB is collaborating with plant breeders and BBSRC to fund research into finding genetic markers that plant breeders can use in their breeding programmes to identify certain traits and use them to develop new varieties,” she says.
One of these projects, MAGIC YIELD, is a BBSRC and AHDB co-funded project in collaboration with NIAB and five plant-breeding companies. Led by Dr James Cockram, crop geneticist at NIAB, the project targets the Holy Grail for plant breeders and farmers, the genetic improvement of grain yield.

Ellie Marshall is hopeful that in the future plant breeders will be able to produce varieties that will deliver in their own right, without over-reliance on chemistry.
“We’re using recently available wheat resources (a multi-parent ‘MAGIC’ population of 1000 lines and high-density genotyping approaches) to investigate the genetic control of yield, yield components, and yield stability.
“The aim is to provide wheat breeders with the molecular tools to help improve the performance of traits in the wheat materials used in their breeding programmes, and ultimately to aid the release of high performing wheat varieties,” he explains.
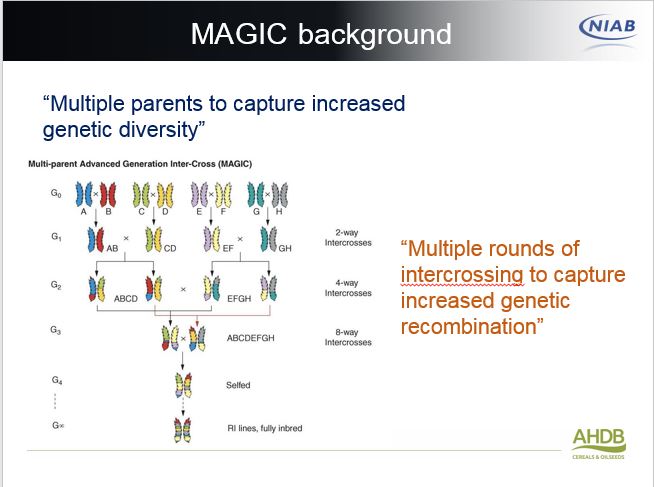
“The MAGIC (Multiparent Advanced Generation Inter-Cross) population of wheat was produced at NIAB by selecting elite wheat varieties and intercrossing them in all combinations to mix up the genetic factors that contribute to yield. After multiple rounds of intercrossing, the resulting progeny were ‘selfed’ to produce 1000 inbred lines.
“Growing and assessing the wheat ‘MAGIC’ population across five locations over two seasons (approx. 5000 2x6m plots in total) has allowed us to identify regions of the wheat chromosomes that control yield and yield components,” reveals James Cockram.
This is done by phenotyping and comparing to genetic marker data. The parent which is contributing to each trait can be tracked and genetic markers developed to help breeders predict traits in their breeding programmes.
Across all 13 traits investigated, a total of approx. 75 chromosomal regions that control their expression have been found.
“We have designed genetic markers for a subset of the most important of these regions. For this subset, we are also investigating the genes and genetic variants we think may be responsible for controlling the traits investigated.
The mapping information and genetic marker production that this project will deliver may provide a platform for more research, believes Keith Gardner, co-investigator in the project.
“We’re providing breeders with signposts rather than precise directions to the exact location of certain traits on the genome. In the near future, we could identify each actual variant in the wheat genome that produces the traits breeders are looking for.
“Because there are so many parents in the MAGIC wheat population, there’s a whole new layer of information to discover. We can more efficiently investigate how locations on the genome interact with one another and whether you can combine these variants for extra resilience,” he suggests.
Background to the MAGIG wheat population
A second wheat-breeding project is under the wing of Dr Paul Nicholson, crop genetics project leader at the John Innes Centre. Its aim is to maximise the potential for Pch1 eyespot resistance and grain protein content in commercial crops of wheat.
Eyespot is a damaging stem base disease that is difficult to control by fungicides and UK losses are estimated to be £12-20M/annum. Two different species cause eyespot so any fungicide must be active against both species to give adequate control.
So what exactly is Pch1? It’s a potent source of eyespot resistance, effective against both eyespot species (Oculimacula yallundae and O. acuformis), but it’s a gene that breeders have been reluctant to employ in varieties because of a perception that it has a negative impact on yield, he explains.
A thumb through the AHDB Recommended List confirms the absence of much substantial eyespot resistance in current varieties, with 35 of the 41 varieties offering ratings of 5 or less. But Pch1 could offer potential benefits over and above just eyespot resistance, because wheat varieties that carry the segment of chromosome with Pch1 also have a higher protein content than expected for their yield.
The project aims to answer a number of questions about Pch1, starting at what exactly is the Pch1 gene? Pch1 was introduced into wheat from an accession (called Vent10) of the wild grass Aegilops ventricosa.
“If the Pch1 gene can be isolated and examined it may give clues to finding more sources of eyespot resistance,” explains Paul Nicholson.
“We have no knowledge at all on the protein effect and don’t know where it is located on the chromosome. Is it separate to the Pch1 eyespot resistance effect? Can lines have eyespot resistance and higher protein without suffering a yield penalty,” he asks.
So what information has the project revealed so far? One of the major revelations is that researchers can find no evidence of any yield impact associated with the gene which should be encouraging news for breeders, he says.
The location of protein effect has also been identified and the good news is that Pch1 along with the region carrying the protein effect is already being introduced into some of the collaborative breeders’ elite varieties, so eyespot resistance and improved protein is already on the way.
The actual location of Pch1 on the chromosome revealed that it’s not the same gene as Pch2, a less potent source of eyespot resistance.
“The major effect Pch1 resistance is in a different part of the chromosome (7D) to the moderate/weak effect Pch2 resistance (7A). It’s not possible to use information from one resistance to isolate/clone the other or look at one resistance to understand more about the other because they’re not the same.
“The region of chromosome containing Pch1 couldn’t be reduced because of lack of recombination. We had to trawl through a large region containing many genes in search of Pch1 itself,” he explains.
Materials provided by the programme of Dr Joseph Jahier of INRA, France have been extremely useful to researchers at JIC and have enabled new angles for addressing some of the many questions surrounding Pch1
“Using Joseph Jahier’s material, we’ve now identified why Pch1 couldn’t be cloned using materials developed over decades. He was years ahead of us in his research and was one of the original team studying Pch1 and his work has been invaluable.
“A real Eureka moment was the realisation that going back to the wild species donor of Pch1 (Aegilops ventricosa) could allow us to examine the chromosomal region containing Pch1 in detail,” adds Paul Nicholson.
Researchers have now developed KASP markers which breeders can use to detect Pch1 in these blind lines, enabling use in breeding programmes. There’s hope that at last breeders will be able to find the needle in the haystack.
The project has now got the point of testing the first Pch1 candidate gene and will have preliminary data within three months.
“While it is extremely challenging to try to clone genes introduced into wheat from wild relatives (due to the fact that there’s little or no recombination after an introgression from the wild relative), it can be done with the appropriate technical and personnel resources.
“If this first gene isolated into wheat does prove to be Pch1, then the experiment wraps up. If it’s not, then we’ll have to go back to the drawing board and revisit genes that don’t seem to behave like resistance genes,” he comments.
But there’s still plenty to learn about Pch1 and how it actually works to defend wheat against eyespot, stresses Paul Nicholson.
“We may be able to identify new sources of eyespot resistance to reduce our reliance on this single potent source and we still haven’t identified the gene responsible for the increased protein effect,” he says.
Perhaps most excitingly is the possibility of increasing the number of copies of Pch1 and thereby increase the eyespot resistance of a variety.
“When you produce an F1 line of wheat, it’s heterozygous for Pch1, meaning it has two different alleles. A homozygous offspring would carry two copies of the same allele. We have shown that homozygous lines are more resistant than heterozygous ones so if we can increase the number of Pch1 copies in a variety then we may see a greater degree of resistance to eyespot infection,” he explains.
“Durum wheat has an A and B genome so if you put Pch1 in position 7A on the chromosome and backcrossed to a hexaploid wheat with Pch1 on 7D, then the offspring will be a new hexaploid wheat with Pch1 on both 7A and 7D.”
These researchers are also looking at whether Pch1 from different lines (vent10 and vent11) can be stacked to produce a wheat more resistant to eyespot.
Summing up, Ellie Marshall describes both projects as steps towards enabling growers to realise the genetic potential of wheat.
“We’re looking more and more towards the plant breeder to produce varieties that will deliver in their own right, without over-reliance on chemistry, by harnessing their innate power,” she concludes.
A plant breeder’s view
Plant breeders are constantly searching for new varieties and Limagrain’s senior wheat breeder, Ed Flatman, believes the two projects will help enhance their breeding programmes.
In the MAGIC yield project, the main focus is productivity, increasing yield through enhanced selection, he explains.
“The eight-parent MAGIC intercross provides an excellent resource to analyse the components of yield, their interaction and the underlying genetics and has a link to our commercial programme, with some of the MAGIC parents being progenitors of many of the Limagrain breeding lines.
“The objective of obtaining accurate markers, by fine mapping the most relevant quantatitive trait loci (QTL), will enable us to actively select for these within our breeding programme using marker assisted selection (MAS),” he continues.
“The data generated within the project will also enable us to develop novel methods such as genomic selection (GS). Both approaches (MAS and GS) will allow us to more rapidly and more precisely accumulate positive genes for yield in new varieties with the objective of increasing the rate of yield gain.”
The value of the Pch1 project is in getting a better understanding of the genetics of eyespot resistance and other traits linked to the resistance gene, highlights Ed Flatman.
“Again an objective is to produce markers for improving the precision of selection for an important trait which is difficult and very laborious to assess on a large scale in a breeding programme.”
In the past, it has been very difficult to utilise Pch1 because of linkage drag of undesirable traits that come with it.
“For some time, there’s been evidence of a yield penalty but also an increase of grain protein content, where the latter is not simply accounted for by a lower yield resulting in an enrichment in protein content.
“If it proves possible to select for the eyespot resistance plus the increased protein content together, without any yield penalty, that would be a real benefit to breeders especially for the selection of bread making quality varieties,” he adds.
Research round-up
AHDB Project No 2200003 MAGIC Map and Go: Deploying MAGIC populations for Rapid Development and Dissemination of Genetic Markers for Yield Improvement in Elite UK Winter Wheat runs from April 2015 to March 2018. The collaborative project between AHDB, NIAB, Bayer CropScience NV, Elsoms Wheat, KWS-UK, Limagrain UK and RAGT UK, is BBSRC LINK sponsored and is costing AHDB £99,544 (10% of project cost). Its aim is to produce genetic markers for breeders to utilise in the quest for yield and yield stability in new varieties.
AHDB Project No 20123803 Maximising the potential for Pch1 eyespot resistance and increased grain protein content in commercial wheat runs from July 2014 to July 2018. It is BBSRC LINK sponsored and a collaboration between AHDB and plant breeders (Biogemma, Limagrain and RAGT) with work carried out at the John Innes Centre at a cost to AHDB of £62,000. The aim is to identify the relative position of genes for three traits (eyespot resistance, yield and protein) and produce plant material to isolate genes for eyespot resistance and protein content from yield. The production of markers will allow breeders to select these desirable traits without unwanted yield effects in their breeding lines.




